

Isolation of Plasmid DNA from E. coli DH5α cells.
Overnight grown culture of E. coli.
A. Luria Broth medium to grow E. coli: Tryptone - 1.0g Yeast extract - 0.5g NaCl - 0.5g Glucose - 0.1g Dissolve the components in 75ml of distilled water, adjust the pH to 7.6 and finally make the volume to 100ml. Sterilize by autoclaving
B. GTE mix: 50mM Glucose 50mM Tris-HCl 10mM EDTA Sterilize by autoclaving
C. 1M NaOH Do not autoclave
D. 10% SDS Dissolve 10g of SDS in 80 ml of distilled water adjust pH to 7.2. Raise the volume to 100ml. Do not Autoclave.
E. Mix II
0.2M NaOH (2.0 ml)
1% SDS (1.0 ml)
DW (7.0ml)
F. 3M Potassium acetate pH 4.8
Potassium acetate 29.4g
DW 25ml
Acetic acid
Adjust pH to 4.8 with acetic acid, raise the volume to 100ml.
All glassware and plastic ware used should be sterilized:
The term 'plasmid' was coined by Joshua Lederberg in 1952, referring to the extrachromosomal DNA of an organism. It is the extrachromosomal DNA of an organism capable of self-replication as well as horizontal transfer, and confers certain characteristics to the bacterium, such as antibiotic resistance, etc. Plasmids are small, circular, double-stranded DNA molecules distinct from a cell's chromosomal DNA. Plasmids naturally exist in bacterial cells, and they also occur in some eukaryotes as well.
(a) Covalently closed circular (CCC) - if both strands are intact circles
(b) Open circular (OC) - if only one strand is intact
(c) Linear (these are exceptions found in some bacteria; e.g., Streptomyces sp.)
Plasmids can be classified as conjugative or non-conjugative depending upon the presence or absence of "tra" genes that permit bacterial conjugation. Also copy numbers of plasmids vary and it also defines the ease with which it can be isolated. Relaxed plasmids have a higher copy number as compared to stringent plasmids which have a limited number. Plasmids encode a few proteins for their own replication and the rest are encoded by the host cell. Replication proteins bind to the sequence called “ori” which is the origin of replication. Purification of plasmid DNA from E.coli culture by alkaline lysis method is based on the principle of differential renaturation of chromosomal and plasmid DNA in order to separate the plasmid DNA and chromosomal DNA. It involves three basic steps:
1. Harvesting and lysis of cells,
2. Separation of plasmid DNA from chromosomal DNA,
3. Purification of plasmid DNA.
Prior to cell lysis, the E. coli cells were treated with Glucose, Tris buffer and EDTA (GTE). EDTA serves dual purpose, it denatures DNases and make cells permeable to sodium dodecyl sulfate (SDS). The GTE solution now protects the bacterial sphaeroplasts so that the membrane bound cells are gently lysed. Then it is treated with alkaline solution which consists of SDS and sodium hydroxide. SDS is an anionic detergent which removes the lipids from the cell membrane and denatures proteins, hence the cell wall becomes weak and the chromosomal DNA and plasmid DNA enters into the solution. Chromosomal DNA is bound to membrane fragments and get separated. Both DNA get denatured at high alkaline pH >12.5. By adding
By adding 3M potassium acetate which acts as neutralizing agent, reducing the pH to 11.5- 12. This helps in the rapid reassociation of both strands of plasmid DNA as it is covalently interwound. The plasmid DNA is purified from the supernatant by alcohol precipitation.
Click here to see animation
Click here to perform the simulation
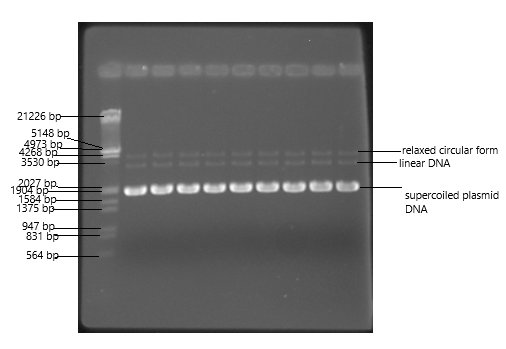
Plasmid DNA may appear in one of the five conformations, which run at different
speeds in a
gel during electrophoresis. The different plasmid conformations are listed below in the order
of electrophoretic mobility.